Mutation decoder
Here is a tool which will allow to translate the mutation information found in genetic reports into a more understandable text.
As of today, the location or the type of mutation is not providing any meaningful information on the impact of syngap1 or on the evolution of the disease.
This tool is only here to provide with better background on what happened during the mutation and to provide more knowledge to parents which are interested.
Introduction
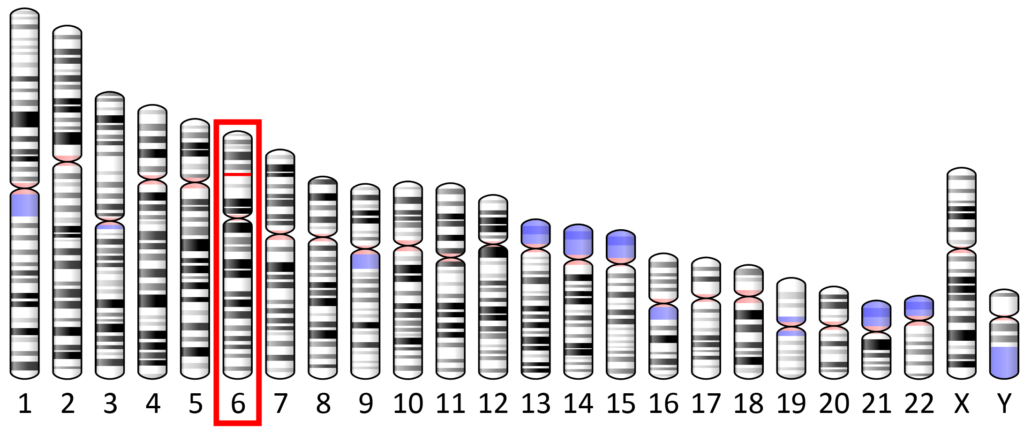
The SYNGAP1 gene is located on Chromosome 6 and is responsible for producing the SynGAP protein. This protein acts as a regulator in the synapses – where neurons communicate with each other. A variant of the SYNGAP1 gene leads to the gene not producing enough SynGAP protein. Without the right amount of SynGAP protein we see an increase in excitability in the synapses making it difficult for neurons to communicate effectively. This leads to many neurological issues seen in SynGAP patients.
Please enter protein notation (start with p.*) or coding DNA notation (start with c.*) in the green box below:
Examples (p.Ser152Cysfs*2 ;
p.Lys835Glnfs10 ; c.3582-4G>A ; c.4373_4377del ; c.1676-1G>C)
Disclaimer
We prioritize the privacy and security of our users. It is essential to clarify that our website does not collect any personal data or information about your device, browser, or online activity. All computations and operations on this platform are performed locally on your computer, ensuring complete confidentiality and control over your data. We do not share any user-specific information with third parties and strictly adhere to all applicable privacy laws. Your trust in us is highly valued, and we remain committed to delivering a safe and secure browsing experience for all our users.
What does it mean ?
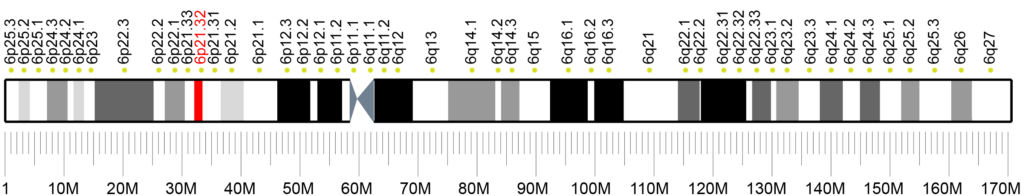
SYNGAP1 Syndrome is caused by a variant on the SYNGAP1 gene (6p.21.32).
The human body is made of trillions of cells. Each cell contains 23 pairs of chromosomes (46 total). Each chromosome contains thousands of genes. Most genes also come in pairs and we get one copy from each parent.
The role of genes is to produce proteins. Proteins are used to regulate the body’s tissues and organs.
A gene can stop working or no longer work properly when a variant (sometimes called a mutation or alteration) occurs. A variant is a mistake that happens, similar to a typo, when the DNA is copied from cell to cell or due to environmental factors.

The 1343 Amino acids composing Syngap1 Protein
MSRSRASIHR GSIPAMSYAP FRDVRGPSMH RTQYVHSPYD RPGWNPRFCI
ISGNQLLMLD EDEIHPLLIR DRRSESSRNK LLRRTVSVPV EGRPHGEHEY
HLGRSRRKSV PGGKQYSMEG APAAPFRPSQ GFLSRRLKSS IKRTKSQPKL
DRTSSFRQIL PRFRSADHDR ARLMQSFKES HSHESLLSPS SAAEALELNL
DEDSIIKPVH SSILGQEFCF EVTTSSGTKC FACRSAAERD KWIENLQRAV
KPNKDNSRRV DNVLKLWIIE ARELPPKKRY YCELCLDDML YARTTSKPRS
ASGDTVFWGE HFEFNNLPAV RALRLHLYRD SDKKRKKDKA GYVGLVTVPV
ATLAGRHFTE QWYPVTLPTG SGGSGGMGSG GGGGSGGGSG GKGKGGCPAV
RLKARYQTMS ILPMELYKEF AEYVTNHYRM LCAVLEPALN VKGKEEVASA
LVHILQSTGK AKDFLSDMAM SEVDRFMERE HLIFRENTLA TKAIEEYMRL
IGQKYLKDAI GEFIRALYES EENCEVDPIK CTASSLAEHQ ANLRMCCELA
LCKVVNSHCV FPRELKEVFA SWRLRCAERG REDIADRLIS ASLFLRFLCP
AIMSPSLFGL MQEYPDEQTS RTLTLIAKVI QNLANFSKFT SKEDFLGFMN
EFLELEWGSM QQFLYEISNL DTLTNSSSFE GYIDLGRELS TLHALLWEVL
PQLSKEALLK LGPLPRLLND ISTALRNPNI QRQPSRQSER PRPQPVVLRG
PSAEMQGYMM RDLNSSIDLQ SFMARGLNSS MDMARLPSPT KEKPPPPPPG
GGKDLFYVSR PPLARSSPAY CTSSSDITEP EQKMLSVNKS VSMLDLQGDG
PGGRLNSSSV SNLAAVGDLL HSSQASLTAA LGLRPAPAGR LSQGSGSSIT
AAGMRLSQMG VTTDGVPAQQ LRIPLSFQNP LFHMAADGPG PPGGHGGGGG
HGPPSSHHHH HHHHHHRGGE PPGDTFAPFH GYSKSEDLSS GVPKPPAASI
LHSHSYSDEF GPSGTDFTRR QLSLQDNLQH MLSPPQITIG PQRPAPSGPG
GGSGGGSGGG GGGQPPPLQR GKSQQLTVSA AQKPRPSSGN LLQSPEPSYG
PARPRQQSLS KEGSIGGSGG SGGGGGGGLK PSITKQHSQT PSTLNPTMPA
SERTVAWVSN MPHLSADIES AHIEREEYKL KEYSKSMDES RLDRVKEYEE
EIHSLKERLH MSNRKLEEYE RRLLSQEEQT SKILMQYQAR LEQSEKRLRQ
QQAEKDSQIK SIIGRLMLVE EELRRDHPAM AEPLPEPKKR LLDAQERQLP
PLGPTNPRVT LAPPWNGLAP PAPPPPPRLQ ITENGEFRNT ADH
Mutation definitions
DNA and RNA
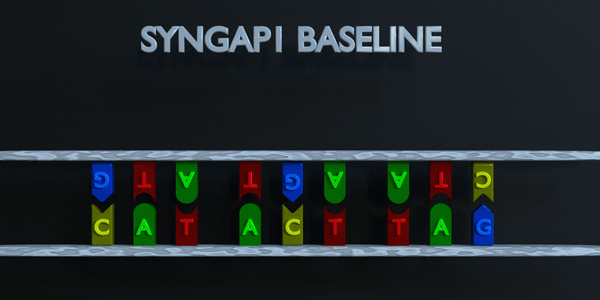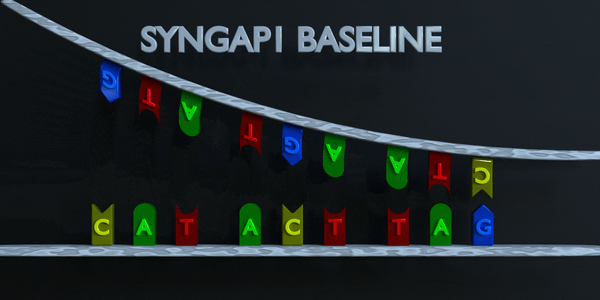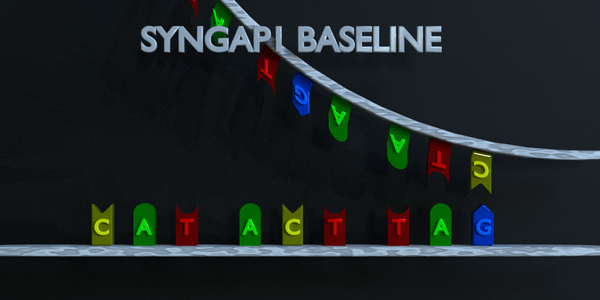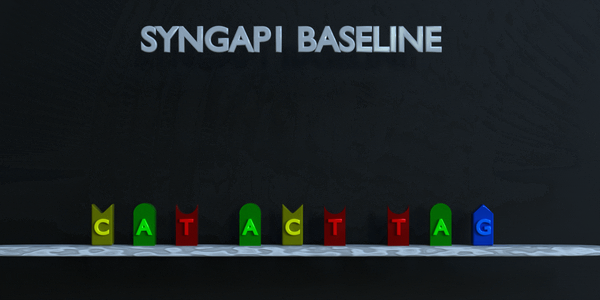
DNA and RNA have a double helix structure. Phosphate groups make up the backbone (in grey), while the middle of the double helix is formed by pairs of nitrogenous bases (colored pieces). Each type of nitrogenous base pairs with another specific base. Cytosine pairs with Guanine, while Adenine pairs with Thymine in DNA, and vice versa. When we talk about RNA (one side of the helix) then Adenine pairs with Uracil. This is why, depending on the notation you read, you may find T (for Thymine) and U (for Uracil). For the sake of simplicity in the explanation below, we will only use the DNA notation. DNA is like a long thread made of “base pairs” A-T and G-C only Genes contain the information to make proteins.
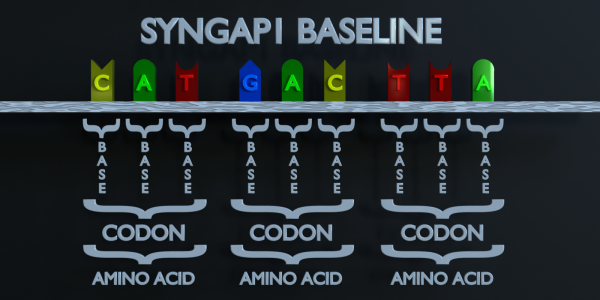
RNA composition
In order for DNA to make proteins, it must be transcribed by messenger RNA (mRNA). The mRNA “reads” the DNA three bases at a time, matching its complementary bases to it. These groups of three bases are called codons, and each codon codes for a different amino acid. Chains of amino acids make up proteins.
One set of three bases means “start”, others specify the 20 different
amino acids (gene building blocks) and some combination means “stop”
The complete list of all bases is called genome
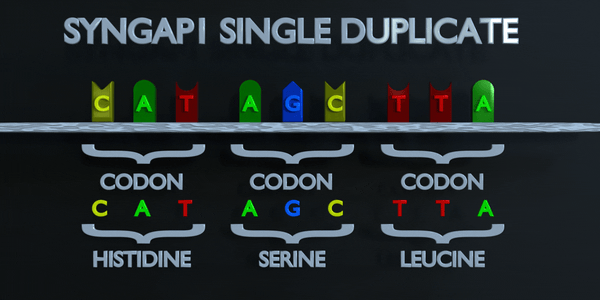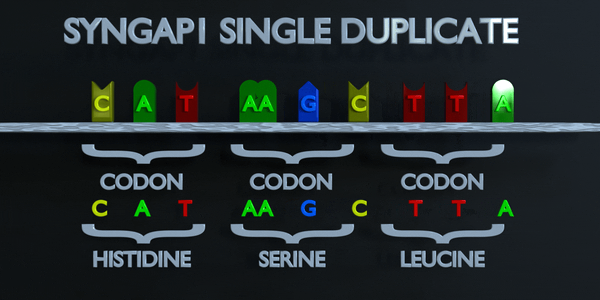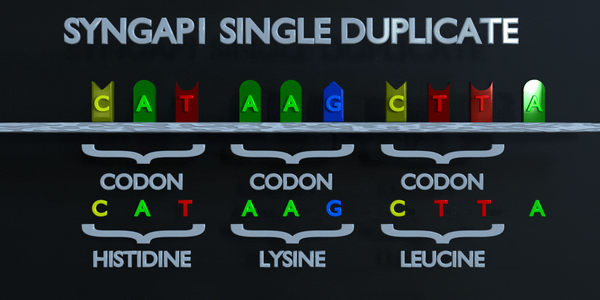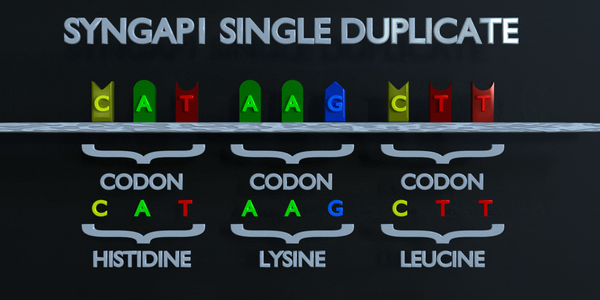
Duplication
As with all mutations, a duplication mutation can drastically change the proteins created by an organism. The proteins responsible for reading DNA process the molecule in units of three base pairs at a time. These codons each specify a different amino acid. If the sequence change even by one nucleotide, a different amino acid is placed within the protein. The function of each protein is dependent on the specific interaction between the amino acids they consist of. A duplicationmutation can displace many more than one nucleotide. In this case, it may make the protein completely dysfunctional, or give it an entirely new function. New adaptations can arise this way, if they are transferred to the offspring and are beneficial. However, the large majority of mutations are deleterious, or cause negative effects.
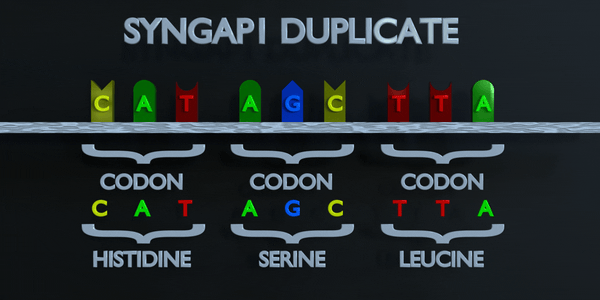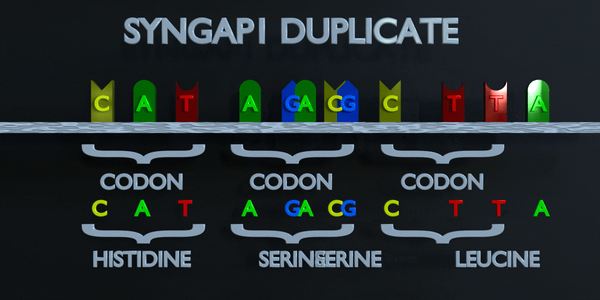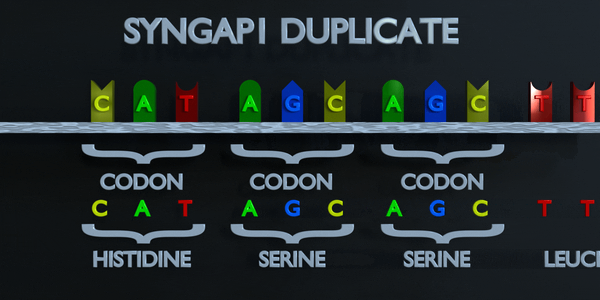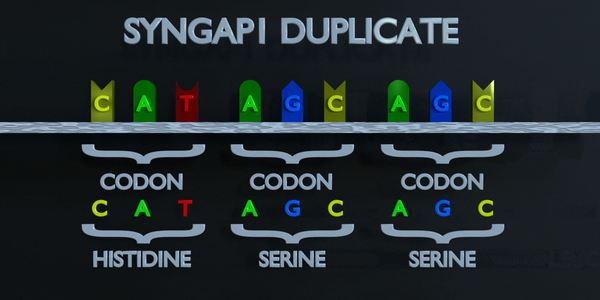
Full Codon Duplication
As with all mutations, a duplication mutation can drastically change the proteins created by an organism. The proteins responsible for reading DNA process the molecule in units of three base pairs at a time. These codons each specify a different amino acid. If the sequence change even by one nucleotide, a different amino acid is placed within the protein. The function of each protein is dependent on the specific interaction between the amino acids they consist of. A duplication mutation can displace many more than one nucleotide. In this case, it may make the protein completely dysfunctional, or give it an entirely new function. New adaptations can arise this way, if they are transferred to the offspring and are beneficial. However, the large majority of mutations are deleterious, or cause negative effects.
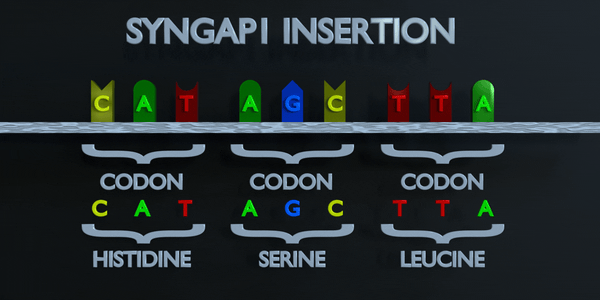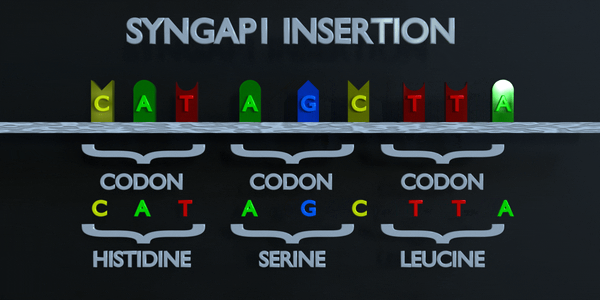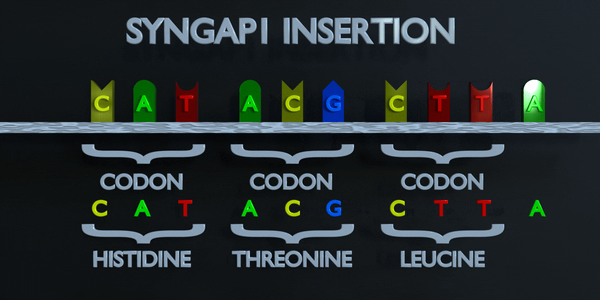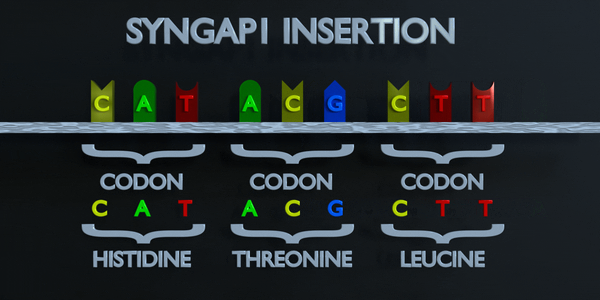
Insertion
Insertions and deletions refer to the addition or removal of short stretches of nucleotide sequences. These types of mutations are usually more deleterious than substitutions since they can cause frame shift mutations, altering the entire amino acid sequence downstream of the mutation site.
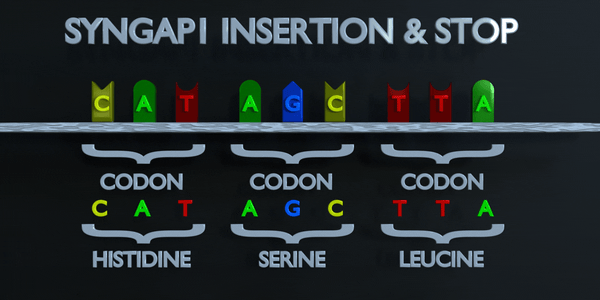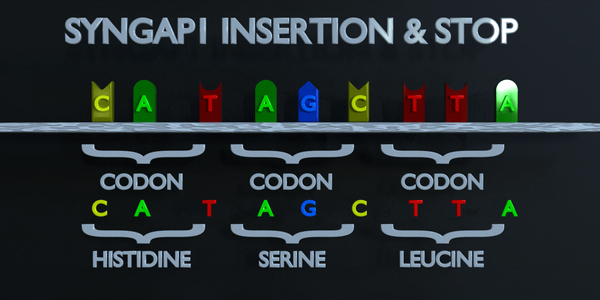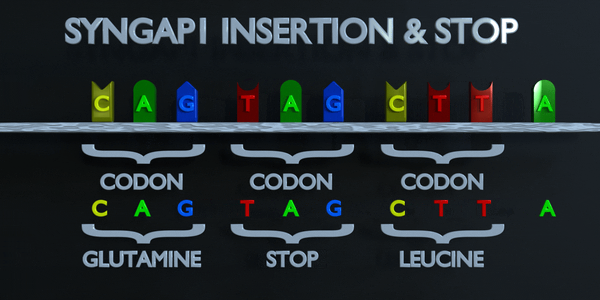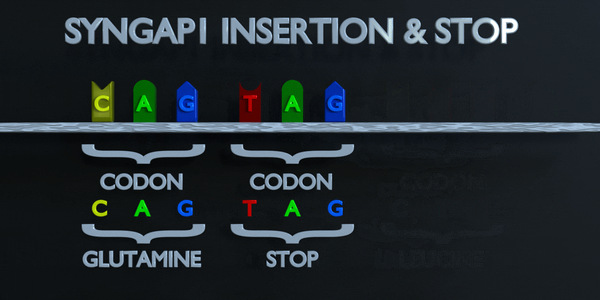
Insertion and Stop (Frameshift)
The genetic code is read in triplets by protein producing enzymes. If three or more nucleotides are added in a gene, entire amino acids can be missing from protein created which can have serious functional effect. Adding a single nucleotide is often not better, as a frameshift mutation can occur. A frameshift mutation shifts the entire gene, and changes all of the original triplet codons. A mutation of this type can cause a gene to produce a completely non-functional gene, as it seriously alters the chain of amino acids the gene produces.
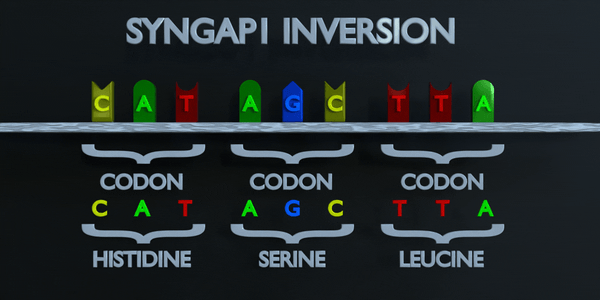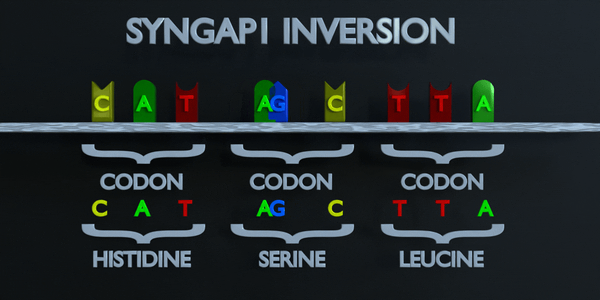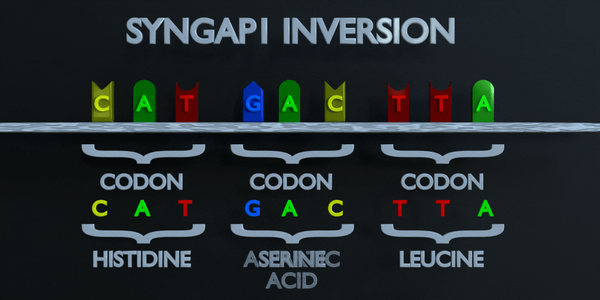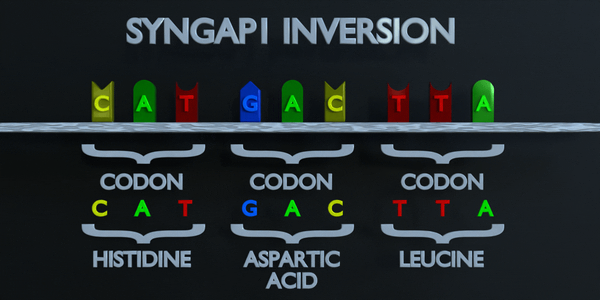
Inversion
Inversion refer to the switching of nucleotide sequences.
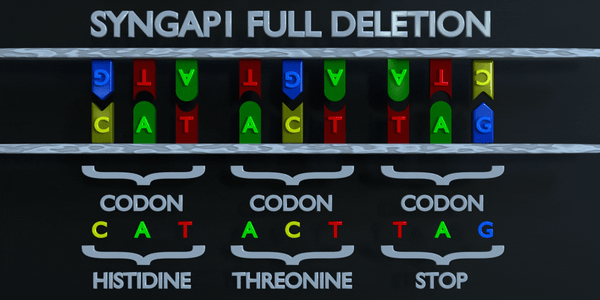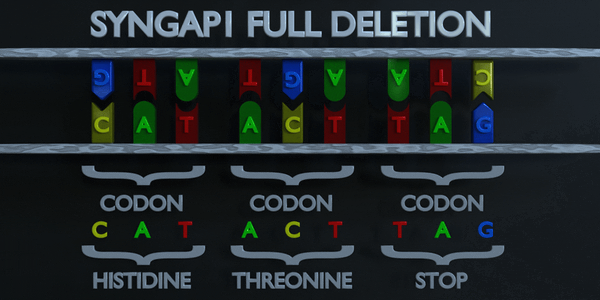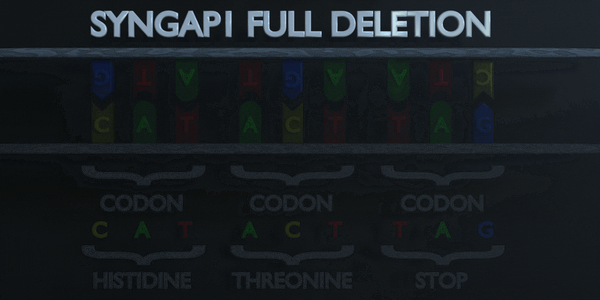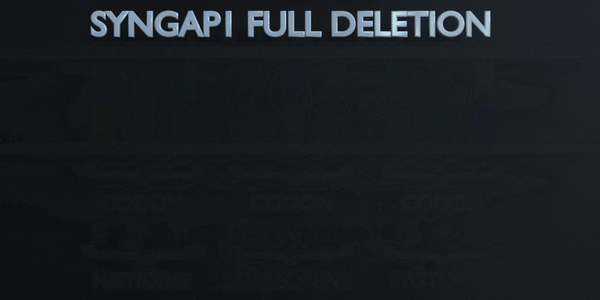
Full Gene Deletion
Sometime, the full gene can be missing (entirely deleted).

Nonsense/Substitution
A nonsense mutation occurs when one nucleotide is substituted and this leads to the formation of a stop codon instead of a codon that codes for an amino acid. A stop codon a certain sequence of bases (TAG, TAA, or TGA in DNA,) that stops the production of the amino acid chain. It is always found at the end of the mRNA sequence when a protein is being produced, but if a substitution causes it to appear in another place, it will prematurely terminate the amino acid sequence and prevent the correct protein from being produced.
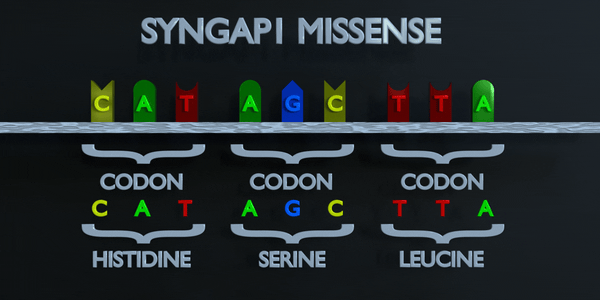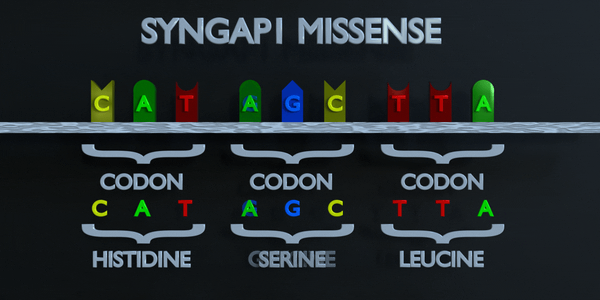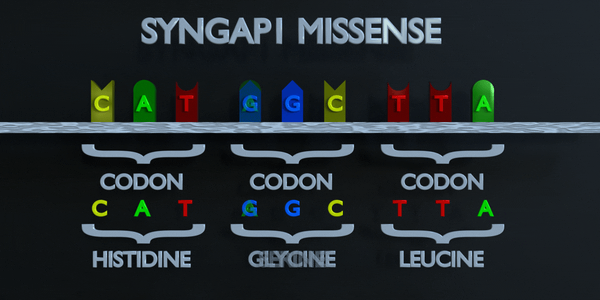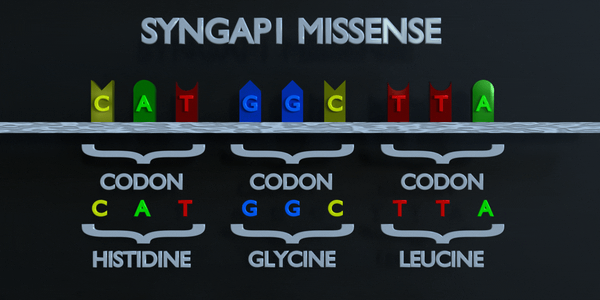
Missense
Like a nonsense mutation, a missense mutation occurs when one nucleotide is substituted and a different codon is formed; but this time, the codon that forms is not a stop codon. Instead, the codon produces a different amino acid in the sequence of amino acids. For example, if a missense substitution changes a codon from AGC to GGC, the amino acid Glycine will be produced instead of Serine. A missense mutation is considered conservative if the amino acid formed via the mutation has similar properties to the one that was supposed to be formed instead. It is called non-conservative if the amino acid has different properties that structure and function of a protein.
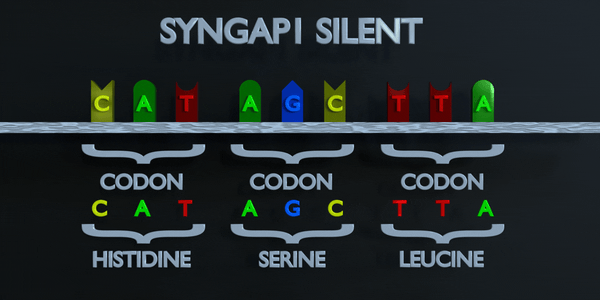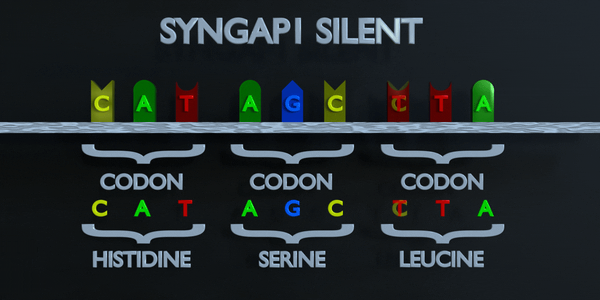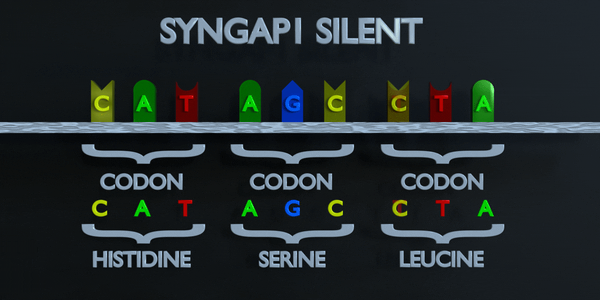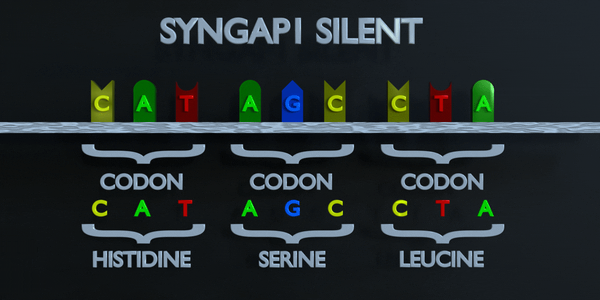
Silent
In a silent mutation, a nucleotide is substituted but the same amino acid is produced anyway. This can occur because multiple codons can code for the same amino acid. For example, TTA and CTA both code for Leucine, so if the first T is changed to a C, the same amino acid will form and the protein will not be affected.
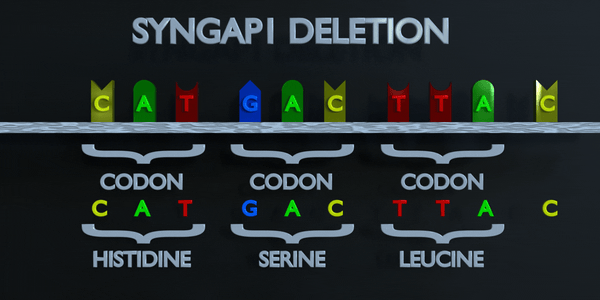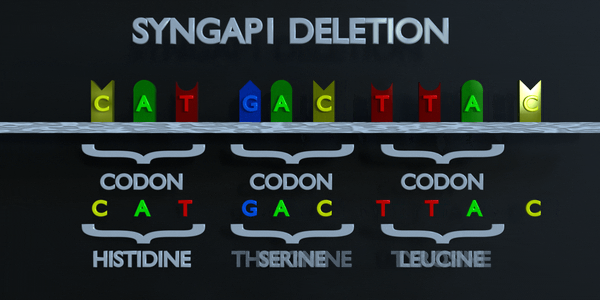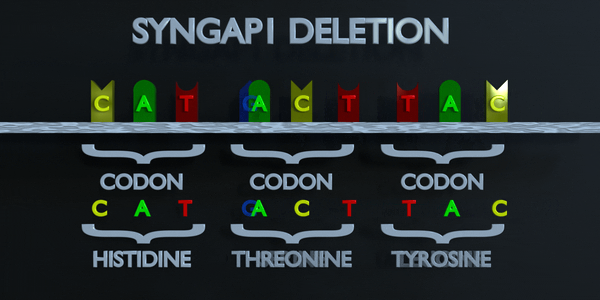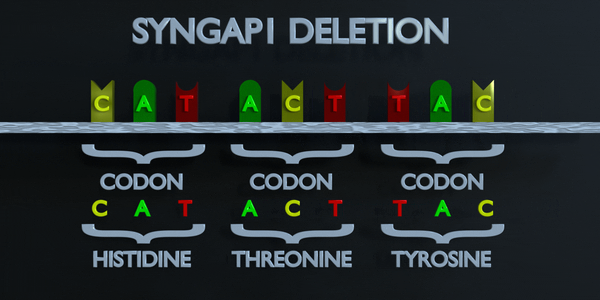
Deletion
A deletion mutation is a mistake in the DNA replication process which removes nucleotides from the genome. A deletion mutation can remove a single nucleotide, or entire sequences of nucleotides. The effect of the deletion mutation will be determined by where in the gene it takes place, and how many nucleotides are deleted. The genetic code is read in triplets by protein producing enzymes. If three or more nucleotides are lost in a gene, entire amino acids can be missing from protein created which can have serious functional effect. Losing a single nucleotide is often not better, as a frameshift mutation can occur. A frameshift mutation shifts the entire gene, and changes all of the original triplet codons. A mutation of this type can cause a gene to produce a completely non-functional gene, as it seriously alters the chain of amino acids the gene produces.